HIGHLIGHTS FROM OUR RESEARCH AT THE DEEPMIA LAB
Deep Learning-based Medical Image Style Translation
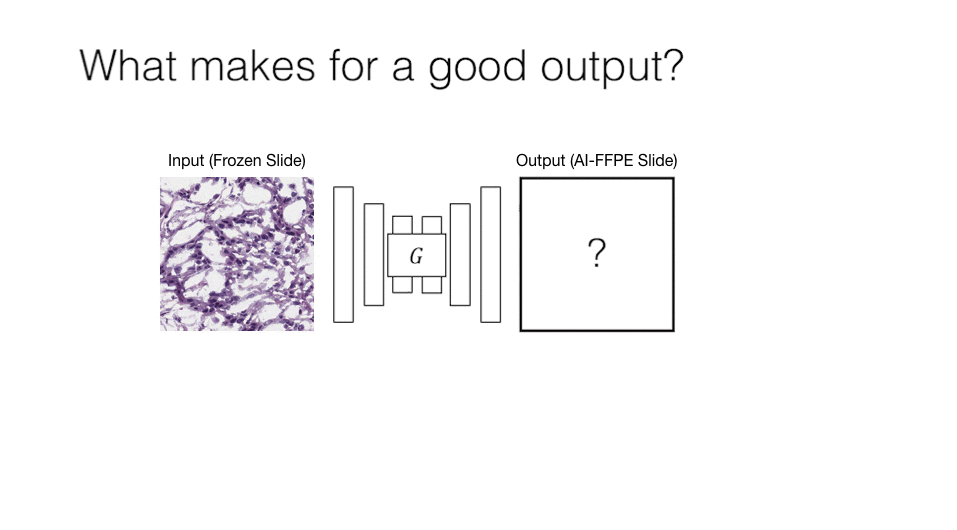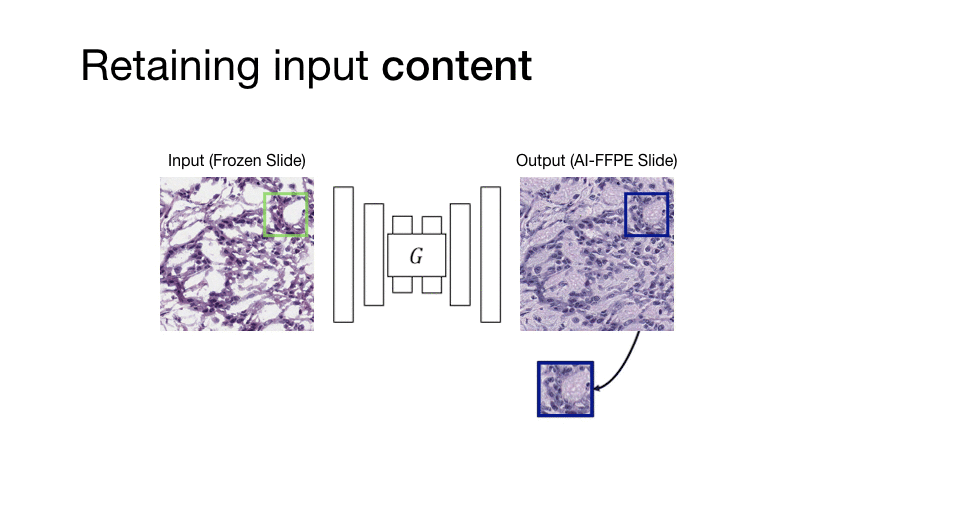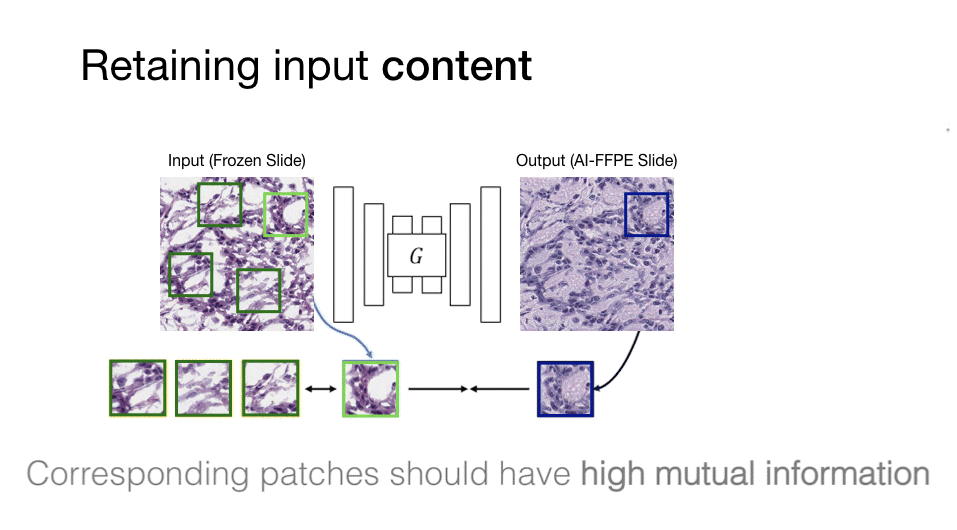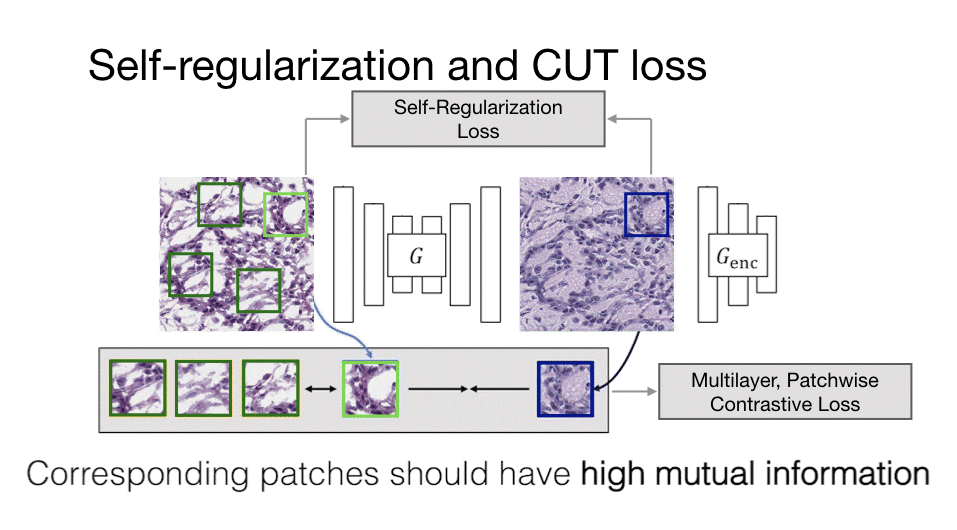
Computational histopathology is a promising field that works on microscopic images to make diagnoses and determine prognostic markers for many diseases. However, staining variation in slides scanned with different tissue types and different scanning devices is one of the critical problems for computational pathology. This staining problem is waiting for a solution as it is an important obstacle for the clinical adaptation of computational pathology solutions. The main goal of our study is to solve this kind of problems with artificial intelligence algorithms, which critically affect the diagnosis in computational histopathology and increase the difficulty of pathologists to make critical decisions in a short time.
EndoSLAM Dataset and an Unsupervised Monocular Visual Odometry and Depth Estimation Approach for Endoscopic Videos
EndoSLAM Dataset Overview

We introduce an endoscopic SLAM dataset which consists of both ex-vivo and synthetically generated data. The ex-vivo part of the dataset includes standard as well as capsule endoscopy recordings. The dataset is divided into 35 sub-datasets. Specifically, 18, 5 and 12 sub-datasets exist for colon, small intestine and stomach respectively.
- To the best of authors' knowledge, this is the very first dataset published to be used in capsule endoscopy SLAM tasks, with timed 6 DoF pose data and high precision 3D map ground truth.
- Two different capsules and conventional endoscope cameras, with high and low resolution were used, so as to generate variety in camera specifications and lighting conditions. Images from different cameras with various resolutions for same organs and depth for each related organs are further unique features of the proposed dataset. We also provide images and pose values for two types of wireless endoscopes, which differ from each other in certain aspects like camera resolution, frame rate, and diagnostic results for detecting Z-line, duodenal papillae and bleeding.
- Some of the sub-datasets include the same trajectories in two versions, e.g with and without polyps so that effect of having polyps as distinguishable features in the organ environment can be analysed, as well.
Sample trajectories from each organ is publicly available in DropBox.
Endo-SfMLearner Overview
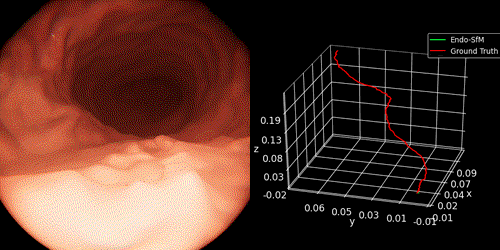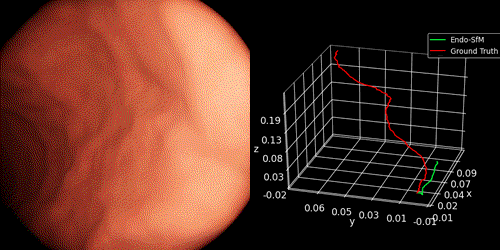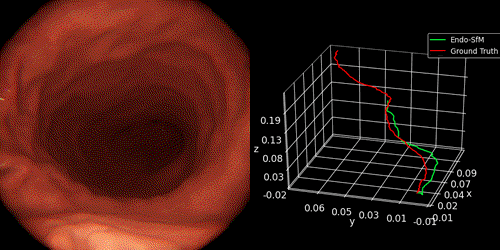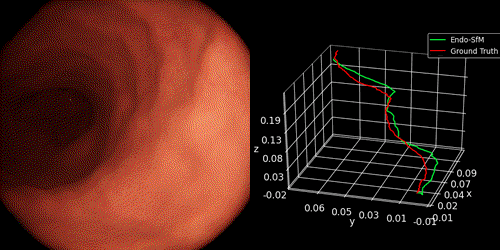
We introduce Endo-SfMLearner framework as self-supervised spatial attantion-based monocular depth and pose estimation method. Our main contributions are as follows:
- Brightness-aware photometric loss, which makes the predicted depth to be consistent under various illumination condition.
- Spatial attention based pose network which is optimized for capsule endoscopy images.
See our GitHub page and our paper for details.
VR-Caps: A Virtual Environment for Capsule Endoscopy
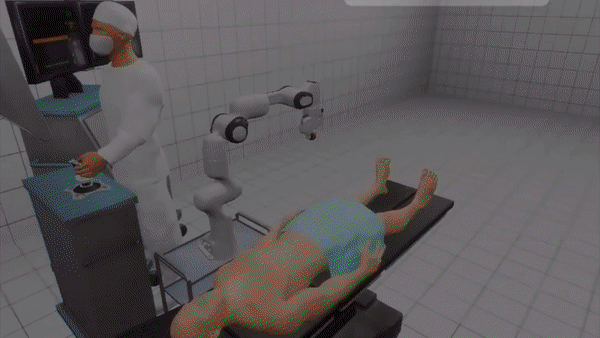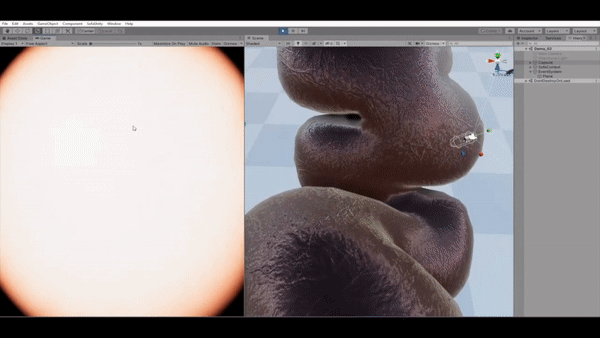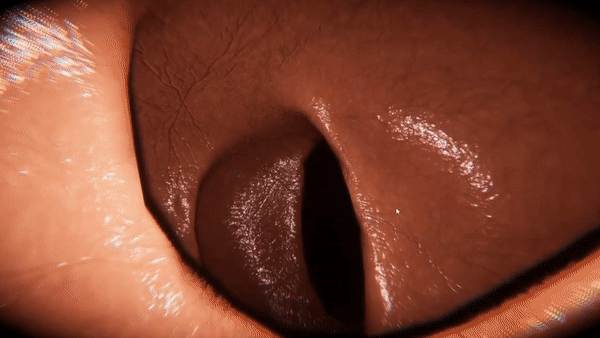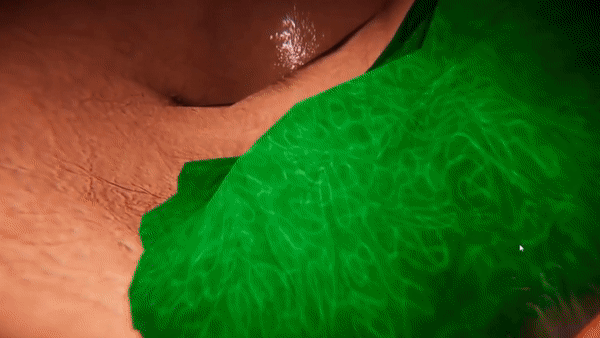
We introduce a virtual active capsule endoscopy environment developed in Unity that provides a simulation platform to generate synthetic data as well as a test bed to develop and test algorithms. Using that environment, we perform various evaluations for common robotics and computer vision tasks of active capsule endoscopy such as classification, pose and depth estimation, area coverage, autonomous navigation, learning control of endoscopic capsule robot with magnetic field inside GI-tract organs, super-resolution, etc. The demonstration of our virtual environment is available on YouTube.
Our main contributions are as follows:
- We propose synthetic data generating tool for creating fully labeled data.
- Using our simulation environment, we provide a platform for testing numerous highly realistic scenarios.
See our GitHub page and our paper for details.
EndoL2H: Deep Super-Resolution for Capsule Endoscopy
We propose and quantitatively validate a novel framework to learn a mapping from low-to-high resolution endoscopic images. We combine conditional adversarial networks with a spatial attention block to improve the resolution by up to factors of 8x, 10x, 12x, respectively. EndoL2H is generally applicable to any endoscopic capsule system and has the potential to improve diagnosis and better harness computational approaches for polyp detection and characterization.
Our main contributions are as follows:
- Spatial Attention-based Super Resolution cGAN: We propose a spatial attention based super-resolution cGAN architecture specifically designed and optimized for capsule endoscopy images.
- High fidelity loss function: We introduce EndoL2H loss which is a weighted hybrid loss function specifically optimized for endoscopic images. It collaboratively combines the strengths of perceptual, content, texture, and pixel-based loss descriptions and improves image quality in terms of pixel values, content, and texture. This combination leads to the maintenance of the image quality even under high scaling factors up to, 10x-12x.
- Qualitative and quantitative study: We conduct a detailed quantitative analysis to assess the effectiveness of our proposed approach and compare it to alternative state-of-the art approaches.
See our GitHub page and our paper for details.


